Computational Methods & Softwares developed/co-developed in our group
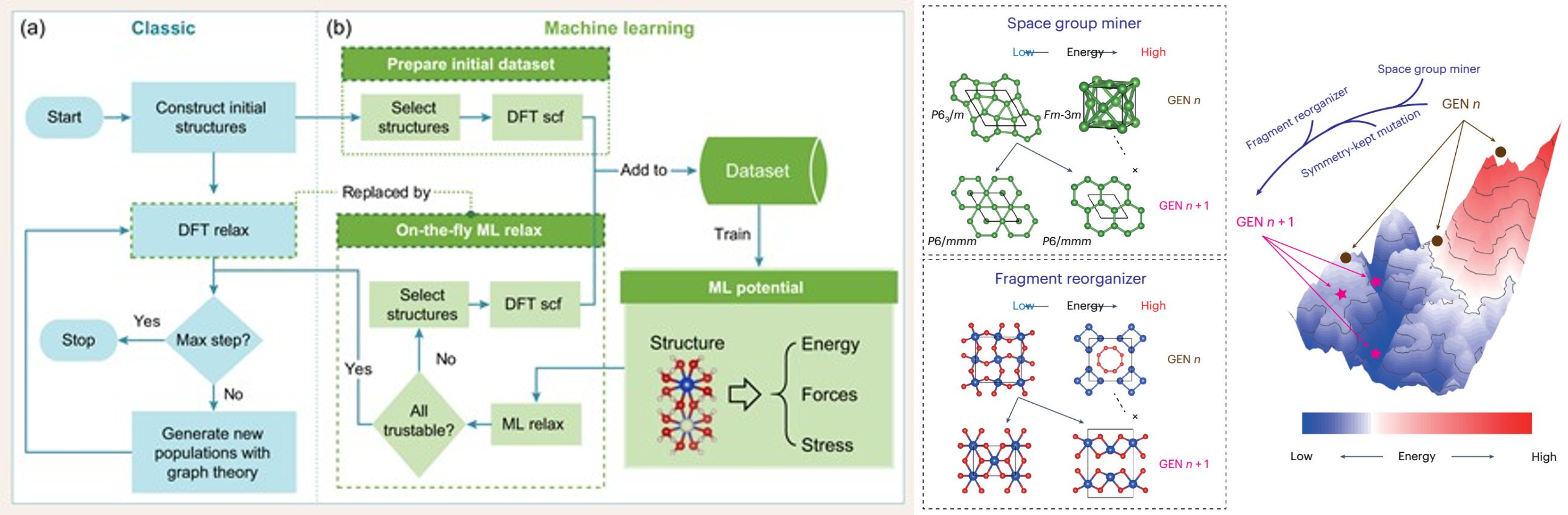
MAGUS: Machine learning and graph theory assisted crystal structure prediction method
Introduction
MAGUS is a machine learning and graph theory assisted crystal
structure prediction method developed by Prof. Jian Sun's group at
the School of Physics at Nanjing University. The programming
languages are mainly Python and C++ and it is built as a pip
installable package. Users can use just a few commands to install
the package. MAGUS has also the advantage of high modularity and
extensibility. All source codes are transparent to users after
installation, and users can modify particular parts according to
their needs. MAGUS has been used to study many systems. Several designed new
materials have been synthesized experimentally, and a number of
high-profile academic papers have been published.
For more details and access to the Magus, visit the code repository.
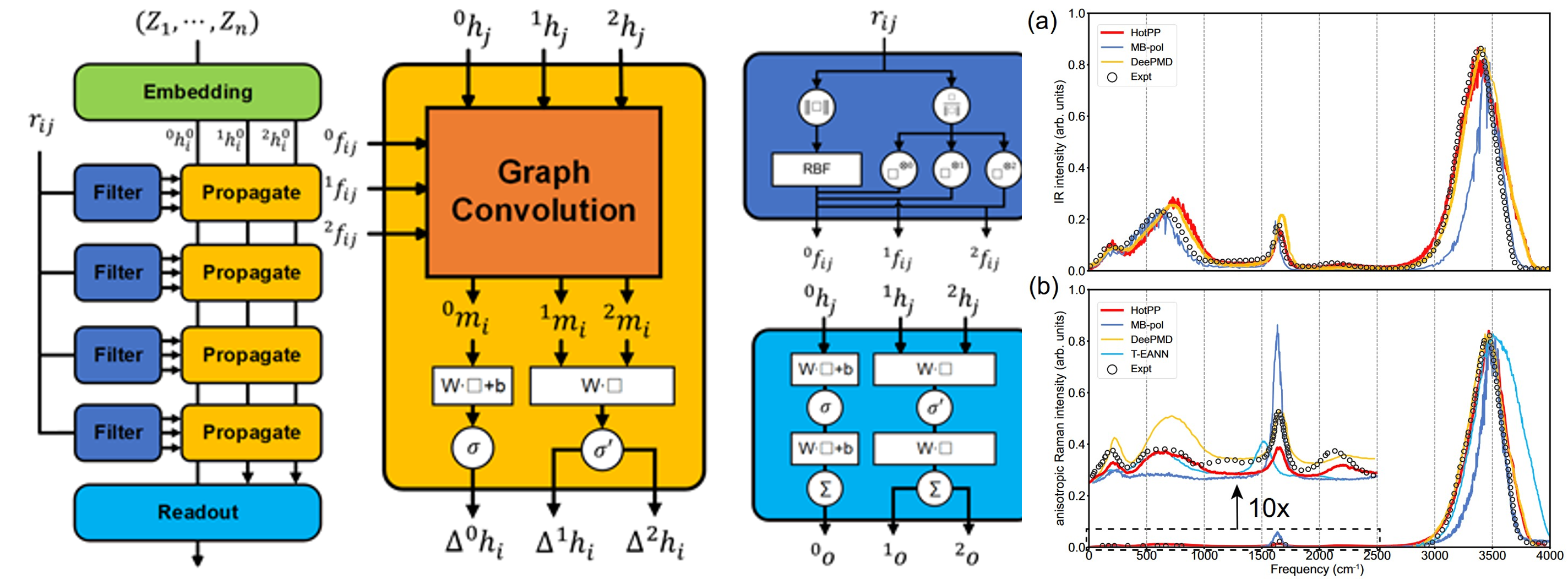
HotPP: High order tensor Passing Potential
Introduction
The higher-order tensor message-passing interatomic potential
function, HotPP, is an E(n)-equivariant message-passing neural
network capable of extending Cartesian tensor embeddings and
messages to arbitrary orders. It supports the calculation of
potential energy surfaces, electric dipole moments, and
polarizabilities. Additionally, it provides interfaces with
commonly used software such as ASE and LAMMPS, making it
applicable for computing phonon spectra, infrared spectra, and
Raman spectra.
For more details and access to the HotPP, visit the code repository.
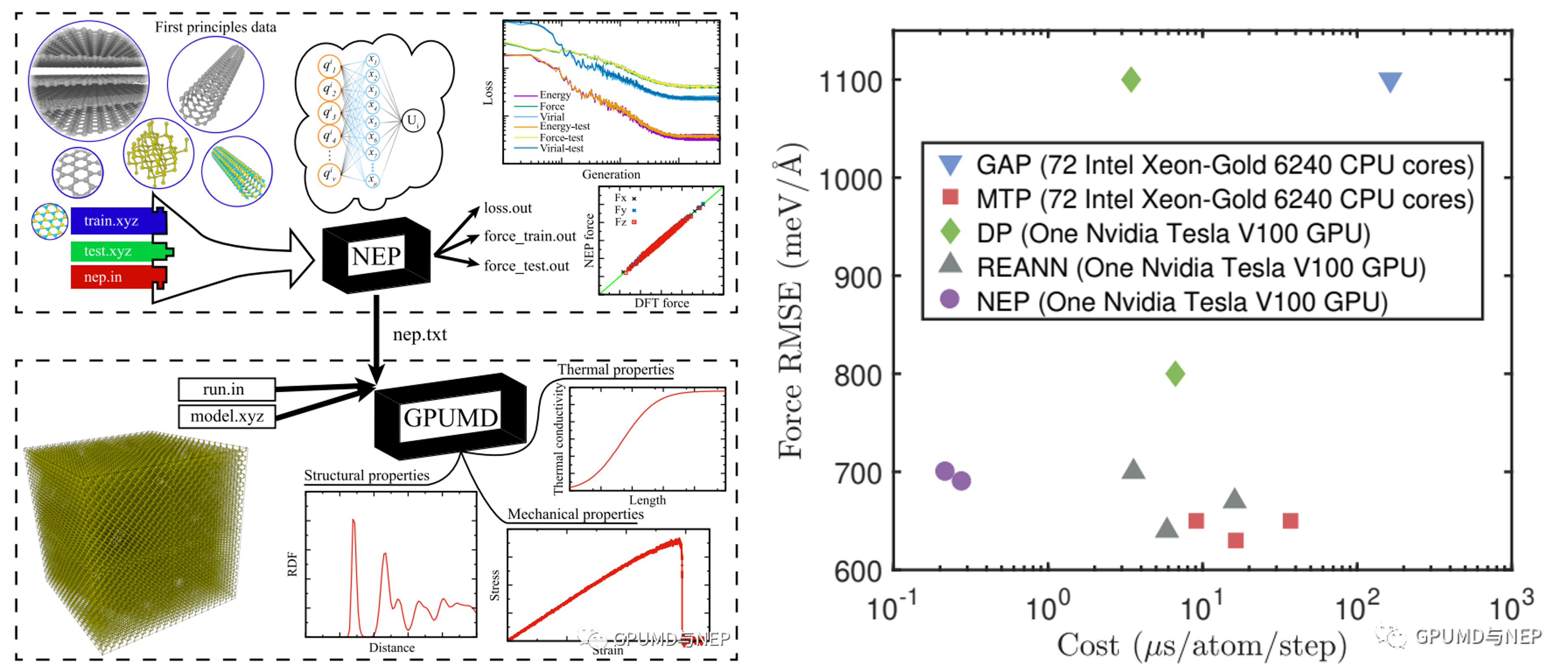
GPUMD: Graphics Processing Units Molecular Dynamics
Introduction
GPUMD (Graphics Processing Units Molecular Dynamics) is a high-performance molecular dynamics simulation software package initially developed and maintained by Professor Zheyong Fan and his team, the first version is released in 2017. At recent years, members from Prof. Jian Sun's group, such as Shuning Pan, Yong Wang, Junjie Wang, Qiuhan Jia, Haoting Zhang, Jiuyang Shi, Zhixing Liang, et al., made a lot of efforts to contribute. Designed to leverage GPU acceleration, it enables efficient simulations of large-scale systems while maintaining high computational accuracy. A key feature of GPUMD is its integration of the Neuroevolution Potential (NEP), a machine-learning interatomic potential trained via evolutionary algorithms, which achieves exceptional computational speed (up to 10^7 atom-steps per second on consumer-grade GPUs) and precision without relying on external ML frameworks like TensorFlow or PyTorch. The software supports diverse functionalities including NVE, NVT, and NPT ensembles, thermal transport analysis (via Green-Kubo, NEMD, and HNEMD methods), structural optimization, phonon dispersion calculations, and elastic constant determination. Compatible with both Windows and Linux systems, GPUMD requires NVIDIA GPUs (compute capability ≥3.5) and CUDA environments for installation. Its ecosystem includes auxiliary tools like PyNEP, GPYUMD, and CALORINE for preprocessing/postprocessing and interoperability with platforms like ASE and LAMMPS. Widely applied in heat transfer, mechanical properties, phase transitions, irradiation damage, and catalysis, GPUMD has been utilized in over 130 high-impact publications and continues to evolve through active community contributions.
For more details and access to the GPUMD, visit the code repository.
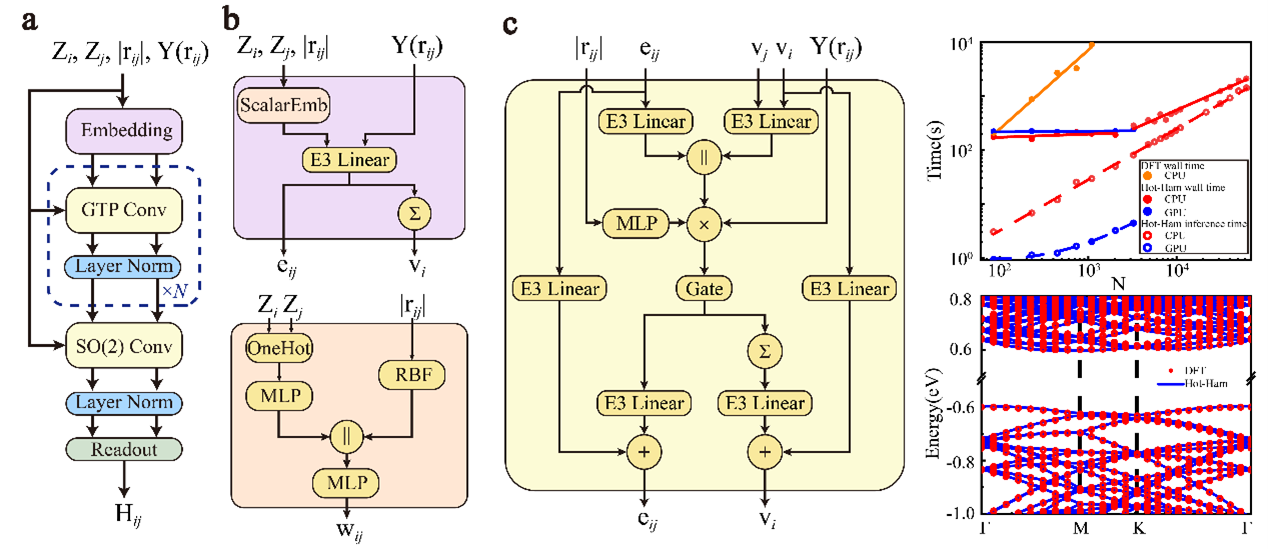
Hot-Ham & GPUTB
Introduction
High-order Tensor machine-learning Hamiltonian (Hot-Ham) is an E(3)-equivariant machine learning models utilizing local coordinate transformation and Gaunt tensor product to achieve efficient high-order spherical tensor products. It enables prediction of density functional theory (DFT) Hamiltonians in the LCAO basis and provides interfaces with ABACUS and OpenMX.
For more details and access to the Hot-Ham, visit the code repository.
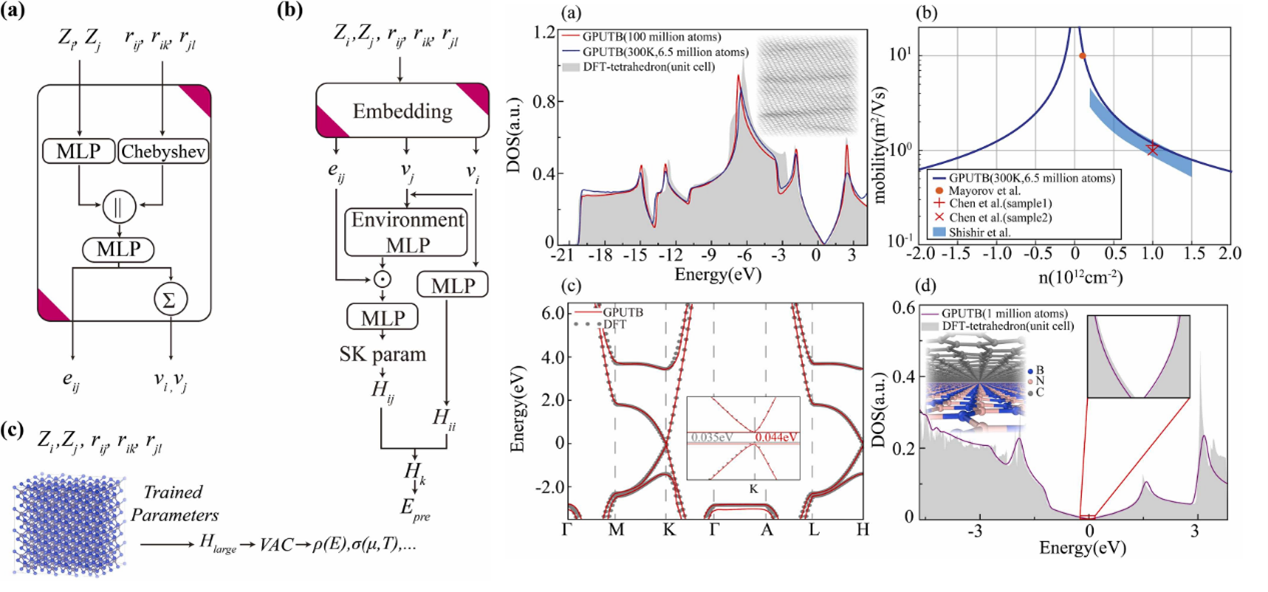
Introduction
GPUTB is a GPU-accelerated machine-learning tight-binding framework that predicts orthogonal LCAO Hamiltonians directly from atomic structures. By using environment-dependent Slater–Koster parameterization and graph neural networks with Chebyshev-based descriptors, GPUTB achieves ab-initio accuracy across different system sizes and temperatures, and enables linear-scaling quantum transport calculations for systems containing millions to hundreds of millions of atoms.
For more details and access to the GPUTB, visit the code repository.